DNA Sequence
Briefly describe the code
Replace on the original sequence
utilize upper() Output upper case results
ending
DNA SequenceACTGATCGATTACGTATAGTATTTGCTATCATACATATATATCGATGCGTTCAT
Seeking complementarity DNA Sequence .
Biologically DNA The brief expression of complementary sequences can be expressed as :A And T,C And G complementary , It can be understood that the existing A use T Instead of ,C use G Instead of ,T use A Instead of ,G use C Instead of , Then the complementary sequence is :
TGACTAGCTAATGCATATCATAAACGATAGTATGTATATATAGCTACGCAAGTA
According to the above statement , I can use replace() Function to replace , take A use T Replace ,T use A Replace ,C use G Replace ,G use C Replace ,
Briefly describe the codemy_dna = "ACTGATCGATTACGTATAGTATTTGCTATCATACATATATATCGATGCGTTCAT"# replace A with Tsequence1 = my_dna.replace('A', 'T')# replace T with Asequence2 = sequence1.replace('T', 'A')# replace C with Gsequence3 = sequence2.replace('C', 'G')# replace G with Csequence4 = sequence3.replace('G', 'C')# print the result of the final replacementprint(sequence1)print(sequence2)print(sequence3)print(sequence4)The output is as follows :
Replace on the original sequenceTCTGTTCGTTTTCGTTTTGTTTTTGCTTTCTTTCTTTTTTTTCGTTGCGTTCTT
ACAGAACGAAAACGAAAAGAAAAAGCAAACAAACAAAAAAAACGAAGCGAACAA
AGAGAAGGAAAAGGAAAAGAAAAAGGAAAGAAAGAAAAAAAAGGAAGGGAAGAA
ACACAACCAAAACCAAAACAAAAACCAAACAAACAAAAAAAACCAACCCAACAA
Obviously the result is incorrect , We are sequence1 To sequence2 An error has already occurred in , Mistake sequence1 in A After being replaced, it becomes T Sequence , stay sequence2 Was replaced in , So we should change our thinking , Keep replacing only the original sequence , No multiple replacements , Avoid mistakes , We can try to replace only the original sequence at a time , Try the following code :
my_dna = "ACTGATCGATTACGTATAGTATTTGCTATCATACATATATATCGATGCGTTCAT"# replace A with Tsequence = my_dna.replace('A', 'T')# replace T with Asequence2 = my_dna.replace('T', 'A')# replace C with Gsequence3 = my_dna.replace('C', 'G')# replace G with Csequence4 = my_dna.replace('G', 'C')print(sequence1)print(sequence2)print(sequence3)print(sequence4)The output is as follows :
TCTGTTCGTTTTCGTTTTGTTTTTGCTTTCTTTCTTTTTTTTCGTTGCGTTCTT
ACAGAACGAAAACGAAAAGAAAAAGCAAACAAACAAAAAAAACGAAGCGAACAA
AGTGATGGATTAGGTATAGTATTTGGTATGATAGATATATATGGATGGGTTGAT
ACTCATCCATTACCTATACTATTTCCTATCATACATATATATCCATCCCTTCAT
Obviously, the result is also incorrect , therefore , We are going to introduce intermediate variables , Finally, make a loop of it ,
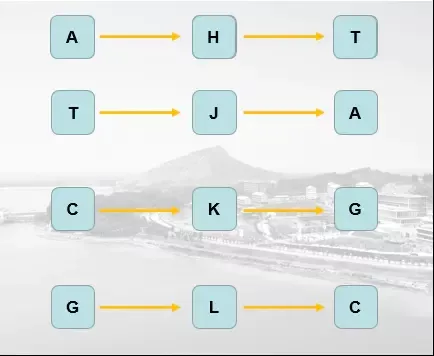
That is to say, four temporary letters are introduced , Then each transform 2 Time , Finally, output the final result , The code can be :
my_dna = "ACTGATCGATTACGTATAGTATTTGCTATCATACATATATATCGATGCGTTCAT"sequence1 = my_dna.replace('A', 'H')sequence2 = sequence1.replace('T', 'J')sequence3 = sequence2.replace('C', 'K')sequence4 = sequence3.replace('G', 'L')sequence5 = sequence4.replace('H', 'T')sequence6 = sequence5.replace('J', 'A')sequence7 = sequence6.replace('K', 'G')sequence8 = sequence7.replace('L', 'C')print(sequence8)The result is :
utilize upper() Output upper case resultsTGACTAGCTAATGCATATCATAAACGATAGTATGTATATATAGCTACGCAAGTA
So far, we have got the result we want , But this method is obviously a little complicated , We can use the case of characters to complete our work , That is, use lowercase letters as temporary variables , In the end use upper() Just output the result in uppercase , The code and results are as follows :
my_dna = "ACTGATCGATTACGTATAGTATTTGCTATCATACATATATATCGATGCGTTCAT"sequence1 = my_dna.replace('A', 't')print(sequence1)sequence2 = sequence1.replace('T', 'a')print(sequence2)sequence3 = sequence2.replace('C', 'g')print(sequence3)sequence4 = sequence3.replace('G', 'c')print(sequence4)print(sequence4.upper())The result is :
tCTGtTCGtTTtCGTtTtGTtTTTGCTtTCtTtCtTtTtTtTCGtTGCGTTCtT
tCaGtaCGtaatCGatatGataaaGCataCtatCtatatataCGtaGCGaaCta
tgaGtagGtaatgGatatGataaaGgatagtatgtatatatagGtaGgGaagta
tgactagctaatgcatatcataaacgatagtatgtatatatagctacgcaagta
TGACTAGCTAATGCATATCATAAACGATAGTATGTATATATAGCTACGCAAGTA
So far, we complement each other DNA The sequence gets , Maybe there's better, simpler code .
endingAlthough this is a small calculation program , But for beginners, every time I upgrade the original code , Even after reading the notes, it feels like a progress , In short, the code is small , The most important thing is to do it ! Hope to learn more Python My fans should not be as ambitious as me , Learning programming is about , reflection , Knock code , reflection , Knock code , Knock code , Re code , More about python seek DNA For information about template complementary sequences, please pay attention to other relevant articles on the software development network !